
Fit ( train_features , train_targets ) # fit the model for training data # predict the 'target' for 'test data' prediction_targets = classifier. If we put 10 then accuracy will be 1.0 # in this example # random_state=23, # keep same proportion of 'target' in test and target data stratify = targets ) # print("Proportion of 'targets' in the dataset") # print("All data:", np.bincount(train_targets) / float(len(train_targets))) # print("Training:", np.bincount(train_targets) / float(len(train_targets))) # print("Training:", np.bincount(test_targets)/ float(len(test_targets))) # use KNeighborsClassifier for classification classifier = KNeighborsClassifier () # training using 'training data' classifier. In below example we can # use train_size=0.4 and test_size=0.2 train_features , test_features , train_targets , test_targets = train_test_split ( features , targets , train_size = 0.8 , test_size = 0.2 , # random but same for all run, also accurancy depends on the # selection of data e.g. Target # both train_size and test_size are defined when we do not want to # use all the data for training and testing e.g. Custom cutoffs can also be supplied as a list of dates to the cutoffs keyword in the crossvalidation function in Python and R.# multiclass_ex.py import numpy as np from sklearn.datasets import load_iris from sklearn.neighbors import KNeighborsClassifier from sklearn.model_selection import train_test_split # create object of class 'load_iris' iris = load_iris () # save features and targets from the 'iris' features , targets = iris. In R, the argument units must be a type accepted by as.difftime, which is weeks or shorter.In Python, the string for initial, period, and horizon should be in the format used by Pandas Timedelta, which accepts units of days or shorter.
Cross Validation Python Code Examples Are
Implements CrossValidation on models and calculating the final result using 'AUCROC method' method. Performs traintestsplit to seperate training and testing dataset 3. Classification metrics used for validation of model. Def crossvalidate(self): Train model using k-fold cross validation and return mean.1. Sum ( prediction_targets = test_targets ) / float ( len ( test_targets )))30 Python code examples are found related to cross validate.
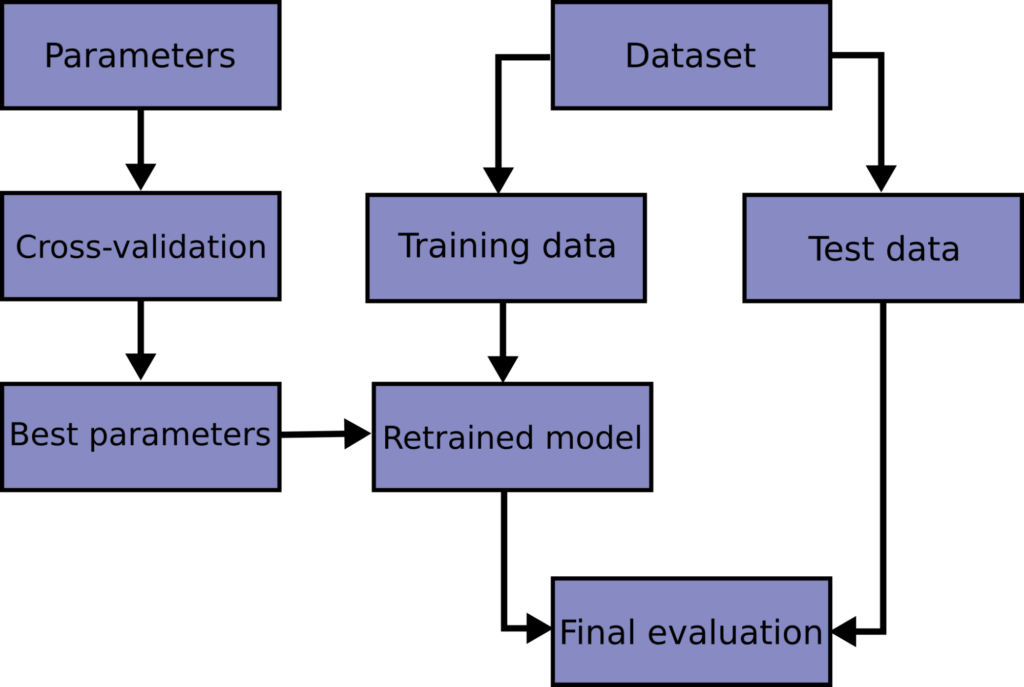
If we put 10 then accuracy will be 1.0 # in this example # random_state=23, # keep same proportion of 'target' in test and target data # stratify=targets # ) # print("Proportion of 'targets' in the dataset") # print("All data:", np.bincount(train_targets) / float(len(train_targets))) # print("Training:", np.bincount(train_targets) / float(len(train_targets))) # print("Training:", np.bincount(test_targets)/ float(len(test_targets))) # use KNeighborsClassifier for classification classifier = KNeighborsClassifier () # training using 'training data' # classifier.fit(train_features, train_targets) # fit the model for training data # predict the 'target' for 'test data' # prediction_targets = classifier.predict(test_features) # check the accuracy of the model # print("Accuracy:", end=' ') # print(np.sum(prediction_targets = test_targets) / float(len(test_targets))) # cross-validation scores = cross_val_score ( classifier , features , targets , cv = 7 ) print ( "Cross validation scores:" , scores ) print ( "Mean score:" , np. In below example we can # use train_size=0.4 and test_size=0.2 # train_features, test_features, train_targets, test_targets = train_test_split( # features, targets, # train_size=0.8, # test_size=0.2, # random but same for all run, also accurancy depends on the # selection of data e.g. Target # both train_size and test_size are defined when we do not want to # use all the data for training and testing e.g. Now execute the code 7 times and we will get different ‘accuracy’ at different run.# multiclass_ex.py import numpy as np from sklearn.datasets import load_iris from sklearn.neighbors import KNeighborsClassifier from sklearn.model_selection import cross_val_score from sklearn.model_selection import train_test_split # create object of class 'load_iris' iris = load_iris () # save features and targets from the 'iris' features , targets = iris. K-fold cross validation splits the data into K-parts, then iteratively use one part for testing and other parts as training data. Python, Supervised Machine Learning / 2 Comments / By Farukh Hashmi.
If we put 10 then accuracy will be 1.0 # in this example # random_state=23, # keep same proportion of 'target' in test and target data # stratify=targets # ) # print("Proportion of 'targets' in the dataset") # print("All data:", np.bincount(train_targets) / float(len(train_targets))) # print("Training:", np.bincount(train_targets) / float(len(train_targets))) # print("Training:", np.bincount(test_targets)/ float(len(test_targets))) # use KNeighborsClassifier for classification classifier = KNeighborsClassifier () # training using 'training data' # classifier.fit(train_features, train_targets) # fit the model for training data # predict the 'target' for 'test data' # prediction_targets = classifier.predict(test_features) # check the accuracy of the model # print("Accuracy:", end=' ') # print(np.sum(prediction_targets = test_targets) / float(len(test_targets))) # print("Targets before shuffle:\n", targets) # rng = np.random.RandomState(0) # permutation = rng.permutation(len(features)) # features, targets = features, targets # print("Targets after shuffle:\n", targets) # cross-validation # cv = KFold(n_splits=3, shuffle=True) # shuffle and divide in 3 equal parts cv = StratifiedKFold ( n_splits = 3 , shuffle = True ) # KFold with 'stratify' option # test_size is available in ShuffleSplit # cv = ShuffleSplit(n_splits=3, test_size=0.2) scores = cross_val_score ( classifier , features , targets , cv = cv ) print ( "Cross validation scores:" , scores ) print ( "Mean score:" , np. In below example we can # use train_size=0.4 and test_size=0.2 # train_features, test_features, train_targets, test_targets = train_test_split( # features, targets, # train_size=0.8, # test_size=0.2, # random but same for all run, also accurancy depends on the # selection of data e.g. Target # both train_size and test_size are defined when we do not want to # use all the data for training and testing e.g. In the iris dataset we have equal number of samples for each target, therefore the effect of shuffle and no-shuffle is almost same, but may vary when targets do not have equal distribution.# multiclass_ex.py import numpy as np from sklearn.datasets import load_iris from sklearn.neighbors import KNeighborsClassifier from sklearn.model_selection import cross_val_score from sklearn.model_selection import train_test_split from sklearn.model_selection import KFold , StratifiedKFold , ShuffleSplit # create object of class 'load_iris' iris = load_iris () # save features and targets from the 'iris' features , targets = iris.



 0 kommentar(er)
0 kommentar(er)
